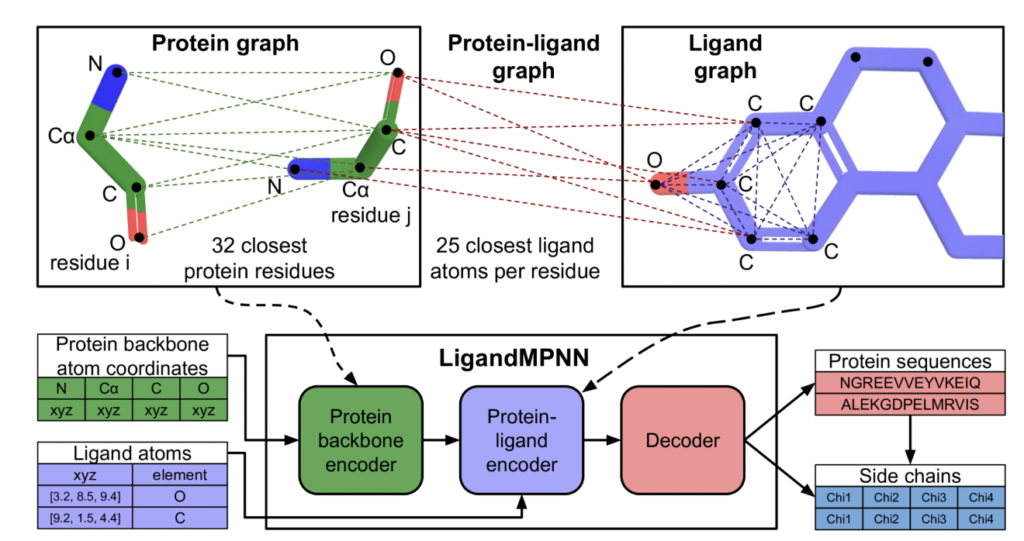
A team of University of Washington researchers have collaborated to address challenges in protein sequence design using LigandMPNN, a deep learning-based protein sequence design method. Model enzymes and target small molecule binders and sensor design. Current physically-based approaches such as Rosetta and deep learning-based models such as ProteinMPNN are unable to accurately model non-protein atoms and molecules, a limitation that hinders the accurate design of protein sequences that contain small molecules, nucleotides and Interact with metals.
The aforementioned methods ignore the obvious consideration of non-protein atoms and molecules, which are important for the design of enzymes, protein-DNA/RNA interactions, and protein-small molecule and protein-metal binders. The proposed solution, LigandMPNN, builds on the ProteinMPNN architecture but explicitly incorporates the full non-protein atom context. LigandMPNN introduces the protein-ligand graph, leveraging neural networks to model interactions and encode ligand atom geometries. This modification leads to LigandMPNN to tailor the sequence and side chain conformation to specific non-protein contexts.
LigandMPNN uses a graph-based approach, treating protein residues as nodes and adding nearest-neighbor edges based on Cα-Cα distances. This model introduces protein-ligand graphs to capture interactions, in which protein residues and ligand atoms represent geometric relationships as nodes and edges. Ligand graphs enhance information transfer in proteins through ligand-protein edges.
Experiments demonstrated better performance of LigandMPNN and its side-chain packing than Rosetta and ProteinMPNN, with 20–30% higher accuracy for small molecules, nucleotides, and metal-interacting residues with high sequence recovery and detailed Demonstrates its effectiveness in structural design. LigandMPNN also outperforms existing models in speed and performance. LigandMPNN is about 250 times faster than Rosetta.
Finally, LigandMPNN fills an important gap in current protein sequence design methods by explicitly incorporating non-protein atoms and molecules. LigandMPNN’s graph-based approach shows significant performance improvements, resulting in high order recovery and high side-chain packing accuracy around small molecules, nucleotides, and metals. LigandMPNN showed exceptional performance in designing small molecule and DNA-binding proteins with high affinity and specificity, which will greatly aid in protein engineering.
check Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us. Twitter And Google News. participation Our 36k+ ML SubReddit, 41k+ Facebook community, Discord channelAnd LinkedIn GrTop.
If you like our work, you will like our work. Newsletter..
Don’t forget to join us. Telegram channel
Pragati Jhanjhanwala is a Consulting Intern at MarketTech Post. She is currently pursuing her B.Tech from Indian Institute of Technology (IIT), Kharagpur. She is a tech enthusiast and has a keen interest in the scope of software and data science applications. She is always reading about the developments in various areas of AI and ML.