× Close
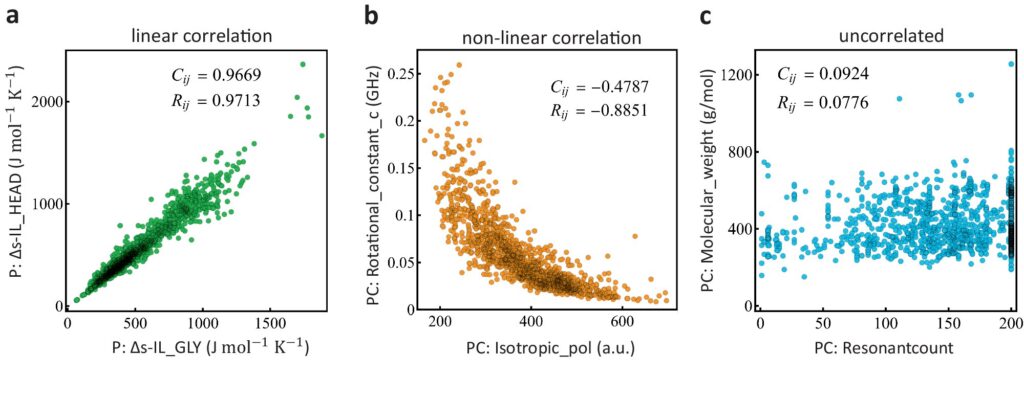
Basic relationships between descriptors. Each data point represents one molecule, and is projected onto a two-dimensional space of two descriptors as shown. The most common features found in descriptors are: a linear correlation, b Non-linear correlations, and c The numbers on each panel of the uncorrelated are calculated from the standardized correlation (i.e., Pearson's coefficient), CIJand rank correlations, RIJ. As shown, rank correlations better capture the non-linear relationships shown in the main panel. Credit: Communication Chemistry (2024). DOI: 10.1038/s42004-024-01161-y
Pathogens are nothing if not adaptive, and their ability to fend off antibiotics is an increasingly public health concern. A research team led by Los Alamos National Laboratory used machine learning, an application of artificial intelligence, to identify molecular features that could lead to the discovery of new types of antibiotics, particularly for these pathogens. which are considered important by the World Health Organization due to their high bacterial count. resistance
The results are published in the journal. Communication Chemistry.
“Some pathogens have properties that make them very effective at resisting antibiotics,” said Ganana Karan, a scientist at Los Alamos. “The discovery of specific compounds capable of inhibiting and propagating certain pathogens is a haystack challenge due to the vast heterogeneity and depth of chemical space, and the complexity of molecular interactions in bacterial membranes. At the pathogen-specific, molecular level profiles that can be tailored for successful drug development.”
Bacterial defense against antibiotics
Gram-negative bacteria have an outer membrane that is less permeable to break down compounds, such as those that make antibiotics, and the bacteria can also excrete compounds that enter, which are antibiotics. reduce the effectiveness of
Data-driven models have the potential to identify molecular features that may overcome such bacterial defenses, but accurate calculations are difficult and consume extensive computing resources. Chemically diverse compounds may contain many related properties. A machine learning-based study narrowed down the relevant spectrum of these properties and established empirical rules that would predict a compound's ability to penetrate the bacterial outer membrane.
Machine learning model identifies pathogen-fighting properties.
Focusing specifically on the Gram-negative bacterium Pseudomonas aeruginosa, the research team developed a machine to identify relevant descriptors associated with compounds and predict the success of these compounds in evading bacterial outer membranes and evasion. Developed a learning model. The team relied on high-performance computing capabilities at Los Alamos to extract the molecular properties of permeation from simulations that considered 1,260 chemically diverse compounds passing through a bacterial membrane.
Their analysis sheds new light on the critical properties that drug candidates need to effectively infect Pseudomonas aeruginosa, and opens the door to similar data-driven studies in other Gram-negative pathogens.
“The machine learning techniques we used in this analysis point to a promising approach for data-driven studies in other biological membranes, including the blood-brain barrier.”
More information:
Pedro D. Manrique et al, Predicting diffusion of compounds across the outer membrane of P. aeruginosa using molecular descriptors, Communication Chemistry (2024). DOI: 10.1038/s42004-024-01161-y
Journal Information:
Communication Chemistry